
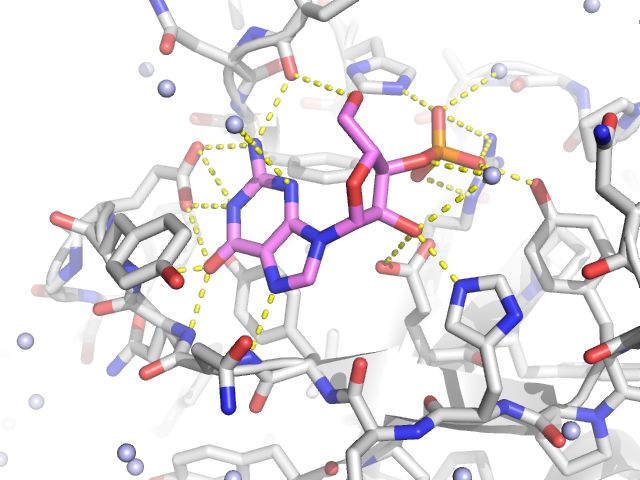
The topology file is very important as it contains all of the forcefield parameters (based upon the forcefield that you select in the initial prompt) for your model.
The p flag is used for output and naming of the topology file. The o flag outputs a new pdb file (various file formats supported) with the name you have given it in the command line. The ff flag is used to select the forcefield (G43a1 is the Gromos 96 FF, a united atom FF). Therefore, we use the ignh flag to ignore hydrogen atoms in the pdb file. This file is derived from an NMR structure which contains hydrogen atoms. The pdb2gmx (to view the command line options for this command just type pdb2gmx h In fact, to get help for any command in Gromacs just use the h flag) command converts your pdb file to a gromacs file and writes the topology for you.
#Pymol tutorial windows#
Text editors in Windows like MS Word add control characters that may cause errors in unix programs.) Process the pdb file with pdb2gmx. In RedHat Linux, use the dos2unix command. to_unix filename filename converts filename to a unix text file. (IMPORTANT! Whenever you sftp a text file to a unix system from Windows, be sure to convert the file to a unix text file. Use sftp to copy your fws.pdb file to the fwspider subdirectories (place a copy in each subdirectory within the fwspider directory). Within this new directory, create the following directories: invacuo, wet, and ionwet. HEADER COMPND FWS TOXIN FWS TOXIN Create a subdirectory in your unix account called fwspider. In addition, delete the SPDBV lines added to the end of the file by DeepView. Edit the file in any text editor and make sure that the HEADER and COMPND line have a name (actually, any name will do!). Use File > Save > Layer and save the new file as fws.pdb.
Deep View fixes the structure and replaces a missing oxygen atom in a proline residue. Open 1OMB.PDB in Deep View (type spdbv 1OMB.pdb in your unix shell). There are no missing side chains in this pdb file, so we will not worry about that in this exercise. DeepView will replace any missing side chains (However, beware as Deep View will mark those rebuilt side chains with a strange control character that can only be 2ģ removed manually using a text editor!). It is advisable to use DeepView (Download DeepView from ) if you know that your structure may be disordered (i.e.
#Pymol tutorial manual#
In addition, you should obtain a copy of the GROMACS user manual at Download 1OMB.PDB from the Protein Data Bank ( We will use MOE to preview the PDB file. To view these files, you must use VMD (Download from: ). We will seek answers to the following questions: Is the secondary structure stable to the dynamics conditions? Are the side chains of the positively charged residues predominantly displaced to one side of the peptide structure? Do the counterions hold these positively charged residues in place or do they move around? What role does water play in maintaining the structure of proteins? Note: You will generate gromacs (*.gro) structure files in this tutorial. We will compare and contrast the impact of counterions in the explicit solvation simulation. We will solvate the peptide in a water box followed by equilibration using Newton s laws of motion. First, we will compare an in vacuo model to a solvated model. We will study this peptide toxin using explicit solvation dynamics. The ion channel is blocked, resulting in blocked nerve signal leading to paralysis and ultimately to death (presumably via respiratory failure). The spider toxin in this tutorial has its positively charged residues pointing predominantly to one side of the peptide. It is hypothesized that exposed positively charged residues in venoms like the spider toxin here bind favorably to the negatively charged entrance of the cell s ion channel. Nerve signals are highly governed by the balance of ions in neuronal cells. Calcium ion channels regulate the entry of this ion into cells. Venom toxins have been used in the past to identify cation channels. In this tutorial, you will study a toxin isolated from the venom of the funnel web spider. You may download it from GROMACS runs on linux, unix, and on Windows (a recent development).
#Pymol tutorial software#
This versatile software package is Gnu public license free software. Biochemistry 32 pp (1993) GROMACS is a high-end, high performance research tool designed for the study of protein dynamics using classical molecular dynamics theory. L.: Sequential assignment and structure determination of spider toxin omega-aga-ivb. AST/IST, University of Medicine and Dentistry of NJ 675 Hoes Lane Piscataway, NJ Phone: (732) Fax: (732)Ģ GROMACS Tutorial for Solvation Study of Spider Toxin Peptide. 1 GROMACS Introductory Tutorial Gromacs ver Author: John E.


 0 kommentar(er)
0 kommentar(er)
